Conformers:
ABBIImiBZICOPNSYNN
Click the Summary/Torsions/Similar/... tabs for more details.
Crystal structure of p19 complexed with 19-bp small interfering RNA
Click a row in table or a step in viewer for analysis of results. Click column headers to sort data.
Steps with non-standard or missing atoms have not been assigned, description of conformers is defined in the table.
Results of the assignment of 36 detected steps in 1 model(s), can be also downloaded as csv or json file. Average confal 0, percentile 0.
Click a row in table or a step in viewer for analysis of results. Click column headers to sort data.
Define restraints for steps within Å cartesian RMSD, global sigma scaling factor or use per step sigma scaling.
| step number | Step name | CANA | NtC | confal | rmsd |
|---|---|---|---|---|---|
| 01 | 1r9f_B_C1_G2 | AAA | AA00 | 89 | 0.26 |
| 02 | 1r9f_B_G2_U3 | AAA | AA00 | 71 | 0.24 |
| 03 | 1r9f_B_U3_A4 | AAA | AA00 | 88 | 0.15 |
| 04 | 1r9f_B_A4_C5 | AAA | AA00 | 86 | 0.22 |
| 05 | 1r9f_B_C5_G6 | AAA | AA00 | 89 | 0.18 |
| 06 | 1r9f_B_G6_C7 | AAA | AA00 | 95 | 0.10 |
| 07 | 1r9f_B_C7_G8 | AAA | AA00 | 89 | 0.13 |
| 08 | 1r9f_B_G8_G9 | AAA | AA00 | 82 | 0.18 |
| 09 | 1r9f_B_G9_A10 | AAA | AA00 | 77 | 0.19 |
| 10 | 1r9f_B_A10_A11 | AAA | AA00 | 73 | 0.15 |
| 11 | 1r9f_B_A11_U12 | AAA | AA08 | 86 | 0.18 |
| 12 | 1r9f_B_U12_A13 | AAA | AA00 | 96 | 0.10 |
| 13 | 1r9f_B_A13_C14 | AAA | AA00 | 97 | 0.11 |
| 14 | 1r9f_B_C14_U15 | AAA | AA00 | 91 | 0.23 |
| 15 | 1r9f_B_U15_U16 | AAA | AA00 | 93 | 0.19 |
| 16 | 1r9f_B_U16_C17 | AAA | AA00 | 94 | 0.17 |
| 17 | 1r9f_B_C17_G18 | AAA | AA00 | 87 | 0.20 |
| 18 | 1r9f_B_G18_A19 | AAA | AA00 | 13 | 0.41 |
| 19 | 1r9f_C_U1_C2 | AAA | AA00 | 89 | 0.19 |
| 20 | 1r9f_C_C2_G3 | AAA | AA00 | 90 | 0.17 |
| 21 | 1r9f_C_G3_A4 | AAA | AA00 | 96 | 0.19 |
| 22 | 1r9f_C_A4_A5 | AAA | AA00 | 95 | 0.11 |
| 23 | 1r9f_C_A5_G6 | AAA | AA00 | 89 | 0.15 |
| 24 | 1r9f_C_G6_U7 | AAA | AA00 | 92 | 0.18 |
| 25 | 1r9f_C_U7_A8 | AAA | AA00 | 92 | 0.17 |
| 26 | 1r9f_C_A8_U9 | AAA | AA00 | 93 | 0.14 |
| 27 | 1r9f_C_U9_U10 | AAA | AA00 | 87 | 0.22 |
| 28 | 1r9f_C_U10_C11 | AAA | AA00 | 86 | 0.20 |
| 29 | 1r9f_C_C11_C12 | AAA | AA00 | 84 | 0.20 |
| 30 | 1r9f_C_C12_G13 | AAA | AA00 | 95 | 0.09 |
| 31 | 1r9f_C_G13_C14 | AAA | AA00 | 95 | 0.07 |
| 32 | 1r9f_C_C14_G15 | AAA | AA00 | 94 | 0.14 |
| 33 | 1r9f_C_G15_U16 | AAA | AA00 | 71 | 0.28 |
| 34 | 1r9f_C_U16_A17 | AAA | AA00 | 86 | 0.21 |
| 35 | 1r9f_C_A17_C18 | AAA | AA00 | 66 | 0.29 |
| 36 | 1r9f_C_C18_G19 | AAA | AA00 | 79 | 0.23 |
Steps with non-standard or missing atoms have not been assigned, description of conformers is defined in the table.
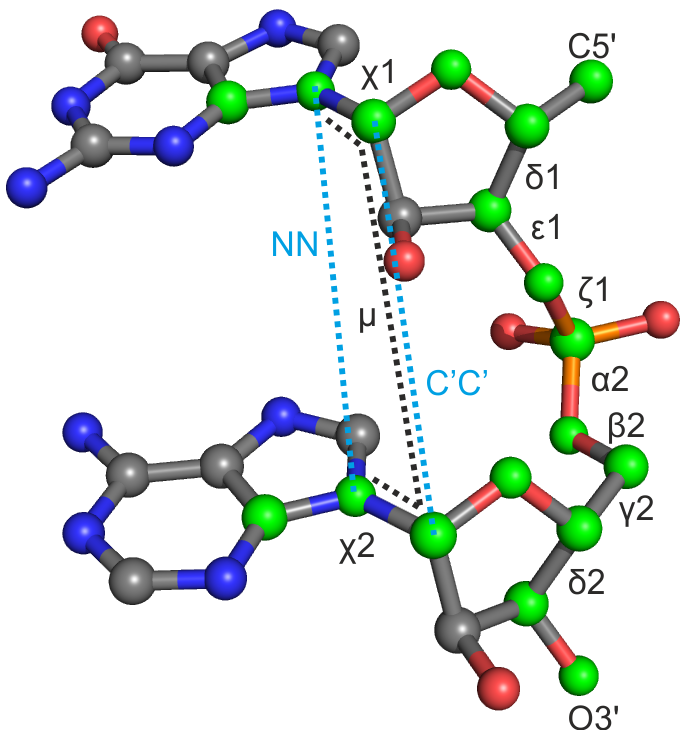
|
step: Table of conformers |
| δ1 | ε1 | ζ1 | α2 | β2 | γ2 | δ2 | χ1 | χ2 | μ | NN | CC | |
| step_torsions | ||||||||||||
| ntC_average | ||||||||||||
| Δ torsions | ||||||||||||
| torsion scores |
| comments | step confal = NaN cartesian RMSD = NaN Å pseudorotation: NA details: NA |
Similarity of step to class averages
Download
Results as csv or json file.Best NtC fitted to input structure.
Restraints
Restraints file for REFMAC (steps with RMSD <= 0.5Å)Commands file for MMB (steps with RMSD <= 0.5Å)
Restraints file for Phenix (steps with RMSD <= 0.5Å) [?]
Definitions
Average parameters (csv), esd values (csv), and Cartesian coordinates of conformers.Download the papers
Description of DNATCO server:Černý et al., Nucleic Acids Research, 44, W284 (2016).
Definition of conformers:
Schneider et al., Acta Cryst D, 74, 52-64 (2018).
Example of application:
Schneider et al., Genes, 8(10), 278, (2017).
Browse conformers
Show
| Color by conformation group (pyramids) | |
|---|---|
| group | |||||||||
| visible |
| Color by NtC (balls) |
|---|
| A | ||||
|---|---|---|---|---|
| A-B | ||||
|---|---|---|---|---|
| B-A | ||||
|---|---|---|---|---|
| B | ||||
|---|---|---|---|---|
| IC | ||||
|---|---|---|---|---|
| OPN | ||||
|---|---|---|---|---|
| Z | ||||
|---|---|---|---|---|
| N | ||||
|---|---|---|---|---|