Conformers:
ABBIImiBZICOPNSYNN
Click the Summary/Torsions/Similar/... tabs for more details.
NMR STRUCTURE OF THE 690 LOOP OF 16 S RRNA OF E. COLI
Click a row in table or a step in viewer for analysis of results. Click column headers to sort data.
Steps with non-standard or missing atoms have not been assigned, description of conformers is defined in the table.
Results of the assignment of 195 detected steps in 15 model(s), can be also downloaded as csv or json file. Average confal 0, percentile 0.
Click a row in table or a step in viewer for analysis of results. Click column headers to sort data.
Define restraints for steps within Å cartesian RMSD, global sigma scaling factor or use per step sigma scaling.
| step number | Step name | CANA | NtC | confal | rmsd |
|---|---|---|---|---|---|
| 001 | 1fhk-m10_A_G1_G2 | AAA | AA00 | 75 | 0.22 |
| 002 | 1fhk-m11_A_G1_G2 | AAA | AA08 | 76 | 0.17 |
| 003 | 1fhk-m12_A_G1_G2 | AAA | AA00 | 85 | 0.15 |
| 004 | 1fhk-m13_A_G1_G2 | AAA | AA00 | 68 | 0.18 |
| 005 | 1fhk-m14_A_G1_G2 | AAA | AA00 | 73 | 0.19 |
| 006 | 1fhk-m15_A_G1_G2 | AAA | AA00 | 73 | 0.22 |
| 007 | 1fhk-m1_A_G1_G2 | AAA | AA00 | 60 | 0.26 |
| 008 | 1fhk-m2_A_G1_G2 | AAA | AA00 | 59 | 0.26 |
| 009 | 1fhk-m3_A_G1_G2 | AAA | AA00 | 64 | 0.23 |
| 010 | 1fhk-m4_A_G1_G2 | AAA | AA00 | 84 | 0.19 |
| 011 | 1fhk-m5_A_G1_G2 | AAA | AA00 | 69 | 0.18 |
| 012 | 1fhk-m6_A_G1_G2 | AAA | AA00 | 63 | 0.22 |
| 013 | 1fhk-m7_A_G1_G2 | AAA | AA08 | 64 | 0.23 |
| 014 | 1fhk-m8_A_G1_G2 | AAA | AA08 | 80 | 0.17 |
| 015 | 1fhk-m9_A_G1_G2 | AAA | AA00 | 51 | 0.19 |
| 016 | 1fhk-m10_A_G2_C3 | AAA | AA00 | 57 | 0.28 |
| 017 | 1fhk-m11_A_G2_C3 | AAA | AA00 | 32 | 0.40 |
| 018 | 1fhk-m12_A_G2_C3 | AAA | AA04 | 88 | 0.28 |
| 019 | 1fhk-m13_A_G2_C3 | AAA | AA00 | 64 | 0.29 |
| 020 | 1fhk-m14_A_G2_C3 | AAA | AA00 | 71 | 0.32 |
| 021 | 1fhk-m15_A_G2_C3 | AAA | AA00 | 56 | 0.30 |
| 022 | 1fhk-m1_A_G2_C3 | AAA | AA00 | 66 | 0.31 |
| 023 | 1fhk-m2_A_G2_C3 | AAA | AA00 | 66 | 0.31 |
| 024 | 1fhk-m3_A_G2_C3 | AAA | AA04 | 86 | 0.16 |
| 025 | 1fhk-m4_A_G2_C3 | AAA | AA00 | 21 | 0.35 |
| 026 | 1fhk-m5_A_G2_C3 | AAA | AA00 | 72 | 0.30 |
| 027 | 1fhk-m6_A_G2_C3 | AAA | AA00 | 66 | 0.27 |
| 028 | 1fhk-m7_A_G2_C3 | AAA | AA00 | 64 | 0.29 |
| 029 | 1fhk-m8_A_G2_C3 | AAA | AA00 | 75 | 0.26 |
| 030 | 1fhk-m9_A_G2_C3 | AAA | AA00 | 67 | 0.30 |
| 031 | 1fhk-m10_A_C3_G4 | NAN | NANT | 0 | 0.48 |
| 032 | 1fhk-m11_A_C3_G4 | AAA | AA04 | 62 | 0.47 |
| 033 | 1fhk-m12_A_C3_G4 | AAA | AA04 | 43 | 0.52 |
| 034 | 1fhk-m13_A_C3_G4 | AAA | AA04 | 42 | 0.52 |
| 035 | 1fhk-m14_A_C3_G4 | AAA | AA04 | 47 | 0.46 |
| 036 | 1fhk-m15_A_C3_G4 | AAA | AA04 | 26 | 0.57 |
| 037 | 1fhk-m1_A_C3_G4 | AAA | AA04 | 51 | 0.49 |
| 038 | 1fhk-m2_A_C3_G4 | AAA | AA04 | 51 | 0.49 |
| 039 | 1fhk-m3_A_C3_G4 | AAA | AA04 | 35 | 0.55 |
| 040 | 1fhk-m4_A_C3_G4 | AAA | AA04 | 47 | 0.54 |
| 041 | 1fhk-m5_A_C3_G4 | AAA | AA04 | 43 | 0.52 |
| 042 | 1fhk-m6_A_C3_G4 | AAA | AA04 | 62 | 0.48 |
| 043 | 1fhk-m7_A_C3_G4 | AAA | AA04 | 50 | 0.56 |
| 044 | 1fhk-m8_A_C3_G4 | AAA | AA00 | 34 | 0.40 |
| 045 | 1fhk-m9_A_C3_G4 | AAA | AA00 | 38 | 0.51 |
| 046 | 1fhk-m10_A_G4_G5 | AAA | AA00 | 62 | 0.22 |
| 047 | 1fhk-m11_A_G4_G5 | AAA | AA00 | 45 | 0.22 |
| 048 | 1fhk-m12_A_G4_G5 | AAA | AA00 | 38 | 0.24 |
| 049 | 1fhk-m13_A_G4_G5 | AAA | AA00 | 24 | 0.32 |
| 050 | 1fhk-m14_A_G4_G5 | AAA | AA00 | 66 | 0.20 |
| 051 | 1fhk-m15_A_G4_G5 | AAA | AA00 | 55 | 0.23 |
| 052 | 1fhk-m1_A_G4_G5 | AAA | AA00 | 59 | 0.19 |
| 053 | 1fhk-m2_A_G4_G5 | AAA | AA00 | 59 | 0.19 |
| 054 | 1fhk-m3_A_G4_G5 | AAA | AA00 | 46 | 0.21 |
| 055 | 1fhk-m4_A_G4_G5 | AAA | AA00 | 23 | 0.25 |
| 056 | 1fhk-m5_A_G4_G5 | AAA | AA00 | 57 | 0.28 |
| 057 | 1fhk-m6_A_G4_G5 | AAA | AA00 | 61 | 0.21 |
| 058 | 1fhk-m7_A_G4_G5 | AAA | AA00 | 65 | 0.23 |
| 059 | 1fhk-m8_A_G4_G5 | AAA | AA00 | 37 | 0.20 |
| 060 | 1fhk-m9_A_G4_G5 | AAA | AA00 | 60 | 0.21 |
| 061 | 1fhk-m10_A_G5_U6 | AAA | AA00 | 39 | 0.38 |
| 062 | 1fhk-m11_A_G5_U6 | AAA | AA00 | 53 | 0.29 |
| 063 | 1fhk-m12_A_G5_U6 | AAA | AA00 | 43 | 0.27 |
| 064 | 1fhk-m13_A_G5_U6 | AAA | AA00 | 18 | 0.33 |
| 065 | 1fhk-m14_A_G5_U6 | AAA | AA00 | 59 | 0.27 |
| 066 | 1fhk-m15_A_G5_U6 | AAA | AA00 | 30 | 0.31 |
| 067 | 1fhk-m1_A_G5_U6 | AAA | AA00 | 46 | 0.26 |
| 068 | 1fhk-m2_A_G5_U6 | AAA | AA00 | 46 | 0.26 |
| 069 | 1fhk-m3_A_G5_U6 | AAA | AA00 | 43 | 0.26 |
| 070 | 1fhk-m4_A_G5_U6 | AAA | AA00 | 47 | 0.28 |
| 071 | 1fhk-m5_A_G5_U6 | AAA | AA00 | 68 | 0.27 |
| 072 | 1fhk-m6_A_G5_U6 | AAA | AA00 | 66 | 0.19 |
| 073 | 1fhk-m7_A_G5_U6 | AAA | AA00 | 42 | 0.26 |
| 074 | 1fhk-m8_A_G5_U6 | AAA | AA00 | 35 | 0.27 |
| 075 | 1fhk-m9_A_G5_U6 | AAA | AA00 | 68 | 0.21 |
| 076 | 1fhk-m10_A_U6_G7 | OPN | OP04 | 40 | 0.25 |
| 077 | 1fhk-m11_A_U6_G7 | OPN | OP04 | 45 | 0.25 |
| 078 | 1fhk-m12_A_U6_G7 | OPN | OP04 | 27 | 0.29 |
| 079 | 1fhk-m13_A_U6_G7 | OPN | OP04 | 26 | 0.29 |
| 080 | 1fhk-m14_A_U6_G7 | OPN | OP04 | 32 | 0.27 |
| 081 | 1fhk-m15_A_U6_G7 | OPN | OP04 | 38 | 0.27 |
| 082 | 1fhk-m1_A_U6_G7 | OPN | OP04 | 44 | 0.23 |
| 083 | 1fhk-m2_A_U6_G7 | OPN | OP04 | 44 | 0.23 |
| 084 | 1fhk-m3_A_U6_G7 | OPN | OP04 | 54 | 0.23 |
| 085 | 1fhk-m4_A_U6_G7 | OPN | OP04 | 61 | 0.22 |
| 086 | 1fhk-m5_A_U6_G7 | OPN | OP04 | 57 | 0.21 |
| 087 | 1fhk-m6_A_U6_G7 | OPN | OP04 | 75 | 0.20 |
| 088 | 1fhk-m7_A_U6_G7 | OPN | OP04 | 63 | 0.24 |
| 089 | 1fhk-m8_A_U6_G7 | OPN | OP04 | 22 | 0.30 |
| 090 | 1fhk-m9_A_U6_G7 | OPN | OP04 | 38 | 0.25 |
| 091 | 1fhk-m10_A_G7_A8 | AAA | AA04 | 57 | 0.42 |
| 092 | 1fhk-m11_A_G7_A8 | AAA | AA00 | 47 | 0.38 |
| 093 | 1fhk-m12_A_G7_A8 | AAA | AA00 | 32 | 0.36 |
| 094 | 1fhk-m13_A_G7_A8 | AAA | AA00 | 25 | 0.37 |
| 095 | 1fhk-m14_A_G7_A8 | AAA | AA00 | 56 | 0.33 |
| 096 | 1fhk-m15_A_G7_A8 | AAA | AA08 | 62 | 0.25 |
| 097 | 1fhk-m1_A_G7_A8 | AAA | AA00 | 27 | 0.39 |
| 098 | 1fhk-m2_A_G7_A8 | AAA | AA00 | 27 | 0.39 |
| 099 | 1fhk-m3_A_G7_A8 | AAA | AA00 | 43 | 0.39 |
| 100 | 1fhk-m4_A_G7_A8 | AAA | AA00 | 70 | 0.32 |
| 101 | 1fhk-m5_A_G7_A8 | AAA | AA00 | 58 | 0.37 |
| 102 | 1fhk-m6_A_G7_A8 | AAA | AA04 | 72 | 0.38 |
| 103 | 1fhk-m7_A_G7_A8 | AAA | AA00 | 68 | 0.34 |
| 104 | 1fhk-m8_A_G7_A8 | AAA | AA00 | 36 | 0.42 |
| 105 | 1fhk-m9_A_G7_A8 | AAA | AA04 | 43 | 0.46 |
| 106 | 1fhk-m10_A_A8_A9 | AAw | AA01 | 37 | 0.39 |
| 107 | 1fhk-m11_A_A8_A9 | AAw | AA01 | 31 | 0.43 |
| 108 | 1fhk-m12_A_A8_A9 | AAw | AA05 | 23 | 0.52 |
| 109 | 1fhk-m13_A_A8_A9 | AAw | AA01 | 45 | 0.35 |
| 110 | 1fhk-m14_A_A8_A9 | AAw | AA01 | 34 | 0.42 |
| 111 | 1fhk-m15_A_A8_A9 | AAw | AA05 | 12 | 0.48 |
| 112 | 1fhk-m1_A_A8_A9 | AAw | AA01 | 24 | 0.40 |
| 113 | 1fhk-m2_A_A8_A9 | AAw | AA01 | 24 | 0.40 |
| 114 | 1fhk-m3_A_A8_A9 | AAw | AA05 | 24 | 0.49 |
| 115 | 1fhk-m4_A_A8_A9 | AAw | AA01 | 39 | 0.38 |
| 116 | 1fhk-m5_A_A8_A9 | AAw | AA01 | 49 | 0.38 |
| 117 | 1fhk-m6_A_A8_A9 | AAw | AA01 | 19 | 0.43 |
| 118 | 1fhk-m7_A_A8_A9 | AAw | AA01 | 69 | 0.30 |
| 119 | 1fhk-m8_A_A8_A9 | AAw | AA01 | 43 | 0.40 |
| 120 | 1fhk-m9_A_A8_A9 | NAN | NANT | 0 | 0.52 |
| 121 | 1fhk-m10_A_A9_A10 | AAA | AA00 | 56 | 0.37 |
| 122 | 1fhk-m11_A_A9_A10 | AAA | AA08 | 60 | 0.29 |
| 123 | 1fhk-m12_A_A9_A10 | AAA | AA00 | 23 | 0.38 |
| 124 | 1fhk-m13_A_A9_A10 | AAA | AA00 | 52 | 0.32 |
| 125 | 1fhk-m14_A_A9_A10 | AAA | AA00 | 41 | 0.34 |
| 126 | 1fhk-m15_A_A9_A10 | AAA | AA08 | 55 | 0.34 |
| 127 | 1fhk-m1_A_A9_A10 | AAA | AA00 | 13 | 0.45 |
| 128 | 1fhk-m2_A_A9_A10 | AAA | AA00 | 13 | 0.45 |
| 129 | 1fhk-m3_A_A9_A10 | AAA | AA00 | 37 | 0.36 |
| 130 | 1fhk-m4_A_A9_A10 | AAA | AA00 | 62 | 0.31 |
| 131 | 1fhk-m5_A_A9_A10 | AAA | AA00 | 50 | 0.39 |
| 132 | 1fhk-m6_A_A9_A10 | AAA | AA00 | 41 | 0.32 |
| 133 | 1fhk-m7_A_A9_A10 | AAA | AA00 | 43 | 0.34 |
| 134 | 1fhk-m8_A_A9_A10 | AAA | AA00 | 47 | 0.34 |
| 135 | 1fhk-m9_A_A9_A10 | AAA | AA00 | 25 | 0.40 |
| 136 | 1fhk-m10_A_A10_U11 | AAA | AA00 | 68 | 0.36 |
| 137 | 1fhk-m11_A_A10_U11 | AAA | AA00 | 62 | 0.26 |
| 138 | 1fhk-m12_A_A10_U11 | AAA | AA00 | 43 | 0.41 |
| 139 | 1fhk-m13_A_A10_U11 | AAA | AA04 | 33 | 0.49 |
| 140 | 1fhk-m14_A_A10_U11 | AAA | AA00 | 87 | 0.22 |
| 141 | 1fhk-m15_A_A10_U11 | AAA | AA00 | 55 | 0.35 |
| 142 | 1fhk-m1_A_A10_U11 | AAA | AA00 | 80 | 0.25 |
| 143 | 1fhk-m2_A_A10_U11 | AAA | AA00 | 80 | 0.25 |
| 144 | 1fhk-m3_A_A10_U11 | AAA | AA00 | 41 | 0.42 |
| 145 | 1fhk-m4_A_A10_U11 | AAA | AA04 | 77 | 0.38 |
| 146 | 1fhk-m5_A_A10_U11 | AAA | AA00 | 44 | 0.35 |
| 147 | 1fhk-m6_A_A10_U11 | AAA | AA00 | 53 | 0.43 |
| 148 | 1fhk-m7_A_A10_U11 | AAA | AA04 | 68 | 0.48 |
| 149 | 1fhk-m8_A_A10_U11 | AAA | AA00 | 87 | 0.21 |
| 150 | 1fhk-m9_A_A10_U11 | AAA | AA00 | 74 | 0.28 |
| 151 | 1fhk-m10_A_U11_G12 | AAA | AA04 | 39 | 0.52 |
| 152 | 1fhk-m11_A_U11_G12 | AAA | AA04 | 16 | 0.56 |
| 153 | 1fhk-m12_A_U11_G12 | AAA | AA04 | 47 | 0.51 |
| 154 | 1fhk-m13_A_U11_G12 | AAA | AA08 | 39 | 0.46 |
| 155 | 1fhk-m14_A_U11_G12 | AAA | AA08 | 40 | 0.44 |
| 156 | 1fhk-m15_A_U11_G12 | AAA | AA04 | 48 | 0.56 |
| 157 | 1fhk-m1_A_U11_G12 | AAA | AA00 | 12 | 0.50 |
| 158 | 1fhk-m2_A_U11_G12 | AAA | AA00 | 12 | 0.50 |
| 159 | 1fhk-m3_A_U11_G12 | AAA | AA04 | 52 | 0.59 |
| 160 | 1fhk-m4_A_U11_G12 | AAA | AA04 | 21 | 0.56 |
| 161 | 1fhk-m5_A_U11_G12 | AAA | AA04 | 40 | 0.49 |
| 162 | 1fhk-m6_A_U11_G12 | AAA | AA04 | 46 | 0.48 |
| 163 | 1fhk-m7_A_U11_G12 | AAA | AA00 | 16 | 0.47 |
| 164 | 1fhk-m8_A_U11_G12 | AAA | AA04 | 32 | 0.54 |
| 165 | 1fhk-m9_A_U11_G12 | AAA | AA04 | 63 | 0.44 |
| 166 | 1fhk-m10_A_G12_C13 | AAA | AA00 | 90 | 0.15 |
| 167 | 1fhk-m11_A_G12_C13 | AAA | AA00 | 75 | 0.24 |
| 168 | 1fhk-m12_A_G12_C13 | AAA | AA00 | 94 | 0.17 |
| 169 | 1fhk-m13_A_G12_C13 | AAA | AA00 | 82 | 0.13 |
| 170 | 1fhk-m14_A_G12_C13 | AAA | AA00 | 77 | 0.23 |
| 171 | 1fhk-m15_A_G12_C13 | AAA | AA00 | 90 | 0.13 |
| 172 | 1fhk-m1_A_G12_C13 | AAA | AA00 | 85 | 0.20 |
| 173 | 1fhk-m2_A_G12_C13 | AAA | AA00 | 85 | 0.20 |
| 174 | 1fhk-m3_A_G12_C13 | AAA | AA00 | 75 | 0.28 |
| 175 | 1fhk-m4_A_G12_C13 | AAA | AA00 | 87 | 0.16 |
| 176 | 1fhk-m5_A_G12_C13 | AAA | AA00 | 77 | 0.17 |
| 177 | 1fhk-m6_A_G12_C13 | AAA | AA00 | 92 | 0.20 |
| 178 | 1fhk-m7_A_G12_C13 | AAA | AA00 | 89 | 0.16 |
| 179 | 1fhk-m8_A_G12_C13 | AAA | AA00 | 80 | 0.24 |
| 180 | 1fhk-m9_A_G12_C13 | AAA | AA00 | 85 | 0.16 |
| 181 | 1fhk-m10_A_C13_C14 | AAA | AA00 | 87 | 0.19 |
| 182 | 1fhk-m11_A_C13_C14 | AAA | AA00 | 75 | 0.31 |
| 183 | 1fhk-m12_A_C13_C14 | AAA | AA00 | 45 | 0.30 |
| 184 | 1fhk-m13_A_C13_C14 | AAA | AA00 | 68 | 0.27 |
| 185 | 1fhk-m14_A_C13_C14 | AAA | AA00 | 78 | 0.27 |
| 186 | 1fhk-m15_A_C13_C14 | AAA | AA08 | 46 | 0.24 |
| 187 | 1fhk-m1_A_C13_C14 | AAA | AA00 | 37 | 0.30 |
| 188 | 1fhk-m2_A_C13_C14 | AAA | AA00 | 37 | 0.30 |
| 189 | 1fhk-m3_A_C13_C14 | AAA | AA00 | 63 | 0.31 |
| 190 | 1fhk-m4_A_C13_C14 | AAA | AA00 | 94 | 0.14 |
| 191 | 1fhk-m5_A_C13_C14 | AAA | AA00 | 42 | 0.44 |
| 192 | 1fhk-m6_A_C13_C14 | AAA | AA00 | 79 | 0.28 |
| 193 | 1fhk-m7_A_C13_C14 | AAA | AA00 | 72 | 0.24 |
| 194 | 1fhk-m8_A_C13_C14 | AAA | AA08 | 56 | 0.27 |
| 195 | 1fhk-m9_A_C13_C14 | AAA | AA00 | 66 | 0.24 |
Steps with non-standard or missing atoms have not been assigned, description of conformers is defined in the table.
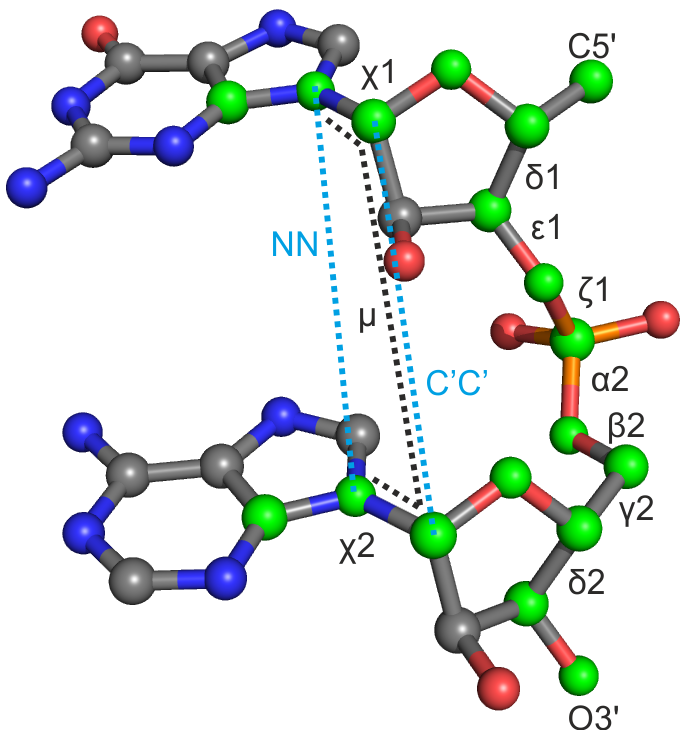
|
step: Table of conformers |
| δ1 | ε1 | ζ1 | α2 | β2 | γ2 | δ2 | χ1 | χ2 | μ | NN | CC | |
| step_torsions | ||||||||||||
| ntC_average | ||||||||||||
| Δ torsions | ||||||||||||
| torsion scores |
| comments | step confal = NaN cartesian RMSD = NaN Å pseudorotation: NA details: NA |
Similarity of step to class averages
Download
Results as csv or json file.Best NtC fitted to input structure.
Restraints
Restraints file for REFMAC (steps with RMSD <= 0.5Å)Commands file for MMB (steps with RMSD <= 0.5Å)
Restraints file for Phenix (steps with RMSD <= 0.5Å) [?]
Definitions
Average parameters (csv), esd values (csv), and Cartesian coordinates of conformers.Download the papers
Description of DNATCO server:Černý et al., Nucleic Acids Research, 44, W284 (2016).
Definition of conformers:
Schneider et al., Acta Cryst D, 74, 52-64 (2018).
Example of application:
Schneider et al., Genes, 8(10), 278, (2017).
Browse conformers
Show
| Color by conformation group (pyramids) | |
|---|---|
| group | |||||||||
| visible |
| Color by NtC (balls) |
|---|
| A | ||||
|---|---|---|---|---|
| A-B | ||||
|---|---|---|---|---|
| B-A | ||||
|---|---|---|---|---|
| B | ||||
|---|---|---|---|---|
| IC | ||||
|---|---|---|---|---|
| OPN | ||||
|---|---|---|---|---|
| Z | ||||
|---|---|---|---|---|
| N | ||||
|---|---|---|---|---|